The Secret in Your Sushi
Posted on Categories Discover Magazine

Photo Credit: Oceana/Jenn Hueting
Dining out or shopping in a grocery store are seemingly straightforward: as the consumer, you make your selection and exchange money for goods. These interactions are based on an implicit trust that you get what you paid for. However, in recent years consumers have begun to demand more transparency with reports of mislabeled seafood at retailers and restaurants being greater than 70% in some instances [1]. Seafood is one of the most traded food items in the world, with approximately 4.5 billion people consuming fish as at least 15% of their source of animal protein [2]. The U.S. is the second largest consumer of seafood in the world behind China and with the recent health recommendations from the American Heart Association elucidating the benefits of fish consumption, sales of this commodity have reached an all-time high [3]. Increased awareness of the environmental burdens of the meat industry have further contributed to this move towards more seafood proteins [4]. The opportunities for seafood mislabeling have consequently increased.
A recent study performed by the Department of Ecology and Evolutionary Biology at UCLA sampled from 26 sushi restaurants in Los Angeles from 2012-2015. Led by Demian A, Willette and Sara E. Simmonds, this study found that a whopping 47% of samples were mislabeled. Similarly, From 2010-2012, Oceana, the world’s largest international ocean conservation organization, conducted a study investigating the prevalence of seafood fraud on a nationwide level. They collected 1,200 samples from 674 restaurants and markets in 21 different states and found that 33% of the samples were mislabeled. Figure 1 depicts a map generated from this study and the respective amount of mislabeling for each state sampled [3].
The types of substitution vary, often substitution cheaper fish such as tilapia for more expensive fish such as grouper, cod, and snapper [3]. Among the different types of fish sampled in UCLA’s four-year study, it was found that all sushi fish types except Bluefin tuna were mislabeled at least once. Halibut and red snapper samples were mislabeled 100% of the time [1].
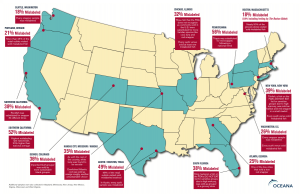
Figure 1: National seafood fraud testing results (Photo Credit: Oceana)
UCLA and Oceana’s studies relied on DNA barcoding technology to elucidate the true identity of the sushi samples. DNA is the genetic blueprint of life, with each organism having its own unique genetic code that can be used in its identification. A DNA barcode is a specific DNA sequence within the genome that is used to identify a species. The sequence chosen is one that is conserved throughout species generations but contains detectable levels of variation between species to serve as a species identifier [5]. The specific sequence used is a fragment of the cytochrome c oxidase I (COI) mitochondrial gene that enables species to be identified without relying on any morphological indicators such as color or shape [4].
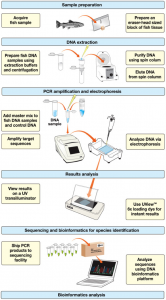
Figure 2: DNA barcoding process. (Photo Credit: Bio-Rad)
The basic process of DNA barcoding is outlined in Figure 2. First, a sample of the fish approximately the size of an eraser on the end of a graphite pencil. Next, the DNA is extracted. This is achieved by the addition of buffers that both aid in breaking the cell membrane (which allows for the release of DNA) and denaturing the DNA [5], which involves the unfolding of its double-stranded helix structure and separation into two single strands, exposing the base pairs and making them available for replication. Now that the fish DNA is isolated, the COI gene must be replicated enough times to be able to be sequenced as well as visualized on a gel. This process is called polymerase chain reaction (PCR) amplification. This is accomplished by using primers, or small strands of DNA that are recognized by DNA polymerase, the enzyme responsible for DNA replication. The primers used are called “degenerate primers,” [5] because they have flexibility in several base positions [6], DNA polymerase is used to extend the primers and replicate the DNA, effectively amplifying the amount of COI gene present in the sample [5].
Figure 3: Scientists prepare fish samples for DNA barcoding. (Photo Credit: Oceana/Jenn Hueting)
A gel electrophoresis is then performed on the sample and a UV transilluminator allows visualization of the DNA banding patterns in each sample. The samples are then sent to a specialized sequencing facility that utilizes the PCR products with a reverse sequencing primer and compares the produced DNA sequences using an algorithm called Basic Local Alignment Search Tool (BLAST) that compares the samples to an established sequence database. This produces a numerical value called the “E-value” that serves as an indicator of homology and is used in conjunction with the result of gel electrophoresis in the identification of each sample’s species [5]. Figure 4 contains an image featuring the DNA barcodes of different species of fish. Each DNA base (C, T, G, or A) is designated by a specific colored bar, which are lined up in sequence and produce a specific barcode. Therefore, color variation indicates which bases differ amongst the species shown [5].
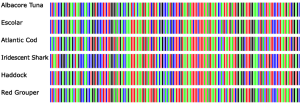
Figure 4: DNA barcodes of different species of fish. (Photo Credit: Ibol Project)
Besides the inherent dishonesty, there are many other negative effects of seafood mislabeling. Ocean conservation programs rely on accurate labeling in their calculations and recommendations, which can be skewed with inaccurate accounts of what species are being caught and sold [1]. Additionally, honest fisherman and businesses suffer as their correctly labeled fish are unable to compete with the low prices of mislabeled fish. Consumer health is also another issue. For example, white tuna was substituted with escolar 84% of the time, which is linked to serious digestive problems and consequently is banned in Italy and Japan [3]. Mislabeling of pufferfish as monkfish has led to temporary neurological damage in some consumers in 2007 and a monkfish recall. Substitution among tuna species also led to elevated mercury levels in canned light tuna, which is usually recommended as a safer canned tuna for children and pregnant women [1].
How can DNA barcoding change the future of seafood mislabeling and the seafood industry? Although 90% of seafood in the U.S. is imported, only 1% is inspected for fraud [3]. If DNA barcoding is used as a regulatory measure, it has the ability to strengthen traceability and therefore liability. Currently it is difficult to pinpoint exactly where in the food chain the mislabeling occurs, whether it is at the restaurant, retailers, or even earlier in the supply chain [3]. By enforcing existing policies through inspectors, retailers, easy-to-use DNA barcoding kits, and a sense of accountability throughout the seafood supply chain, we can use science to move towards a resolution towards these fishy mislabeling practices.
References Cited:
- Willette, D.A., Simmonds, A.E., Cheng, S.H., et. Al. (2017). Using DNA barcoding to track seafood mislabeling in Los Angeles restaurants. doi: 10.1111/coni.12888.
- Bene, C., Barange, M., Subasinghe, R. et al. (2015). Feeding 9 billion people by 2050 – putting fish back on the menu. Food Security 7: 261-274.
- Oceana (2013). Oceana study reveals seafood fraud nationwide. Retrieved from http://oceana.org/sites/default/files/National_Seafood_Fraud_Testing_Results_Highlights_FINAL.pdf
- Wong, E.H.K., Hanner, R.H. (2008). DNA barcoding detects market substitution in North American seafood. Food Research International 41: 828-837.
- Tighe, D., Andrews, S., Brown, L. (2016). Generate a DNA barcode and identify species. Retrieved from http://slideplayer.com/slide/5768597/
- Linhart, C., Shamir, R. (2005). The degenerate primer design problem: theory and applications. J comput Biol 4: 431-56.
 About the author: Ashton Yoon received her B.S. in Environmental Science at UCLA and is currently pursuing a graduate degree in food science. Her favorite pastime is experimenting in the kitchen with new recipes and cooking techniques.
About the author: Ashton Yoon received her B.S. in Environmental Science at UCLA and is currently pursuing a graduate degree in food science. Her favorite pastime is experimenting in the kitchen with new recipes and cooking techniques.