Interbreeding Surprise! More Denisovans In Our Family Tree
Posted on Categories Discover Magazine
Genetic analysis reveals new evidence of Homo sapiens interbreeding with Denisovans, an extinct species closely related to Neanderthals and known from a handful of fossils, such as this toe bone. (Credit Bence Viola/Wikimedia Commons)
Hey, sex happens. And apparently, whenever our ancestors met up with other members of the genus Homo, it happened a lot. New genetic analysis published today reveals previously unidentified evidence of interbreeding between Homo sapiens and Denisovans, a branch of our family tree not even known to science until a decade ago.
You remember the Denisovans, right? Researchers uncovered a piece of pinky finger and a few other fossil fragments in Denisova Cave, in the Altai Mountains of Siberia, in the late ’00s. The full significance of the discovery wasn’t really appreciated until 2010, however, when genetic analysis revealed that the bones belonged not to H. sapiens or Neanderthals, both of whom frequented the site, but to a completely new species of hominin.
Since that discovery, teams have been combing through DNA from both ancient samples and modern populations, hunting for evidence of interbreeding between the Denisovans and our own species. Researchers already have significant data that H. sapiens interbred with Neanderthals multiple times, so it seemed a good bet that there was also a bit of hominin hanky-panky between us and the Denisovans.
And, lo and behold, studies did uncover evidence of Denisovan ancestry in modern H. sapiens populations, in certain parts of Asia, especially Oceania, including Indonesia’s Papua province, where locals’ DNA was up to about 5 percent Denisovan-derived. (For comparison, about 2 percent of the DNA of most non-Africans alive today comes courtesy of past Neanderthal interbreeding events.)
In 2014, Nature published a study that found a variation of a gene, which regulates hemoglobin and is present in modern Tibetans, was inherited from Denisovans. The genetic hand-me-down is likely what made it possible for people to live at 13,000 feet or more above sea level in the oxygen-sparse air of the Himalayas.
Today, however, applying a new data-modeling method to more than 5,600 genomes from modern populations, researchers identified surprising new details about interbreeding between H. sapiens and Denisovans: It appears different populations of the two hominins interbred multiple times.
In parts of China and Japan, people inherited most of their Denisovan DNA from one population of the archaic humans, but Papuans in Oceania got theirs from a different Denisovan lineage.
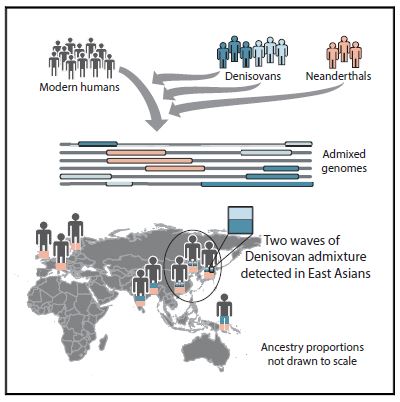
Researchers uncovered genetic evidence of at least two separate interbreeding events between Denisovans and Homo sapiens.
(Credit Browning et al)
Mixing It Up
What’s particularly interesting about this unexpected discovery is that the Denisovan-derived DNA seen in modern Papuans stands out even more than it did before. To understand why, consider that the amount of Denisovan DNA in modern Chinese and Japanese genomes included in the study is only about 0.2 percent, significantly less than the roughly 5 percent of the Denisovan contribution to modern Papuans. That 0.2 percent of Denisovan-derived DNA in East Asian genomes turns out to be closely related to the original Altai Denisovan genome sequenced in 2010.
(Sidenote: Although it’s closely related, researchers can’t determine whether the 2010 genome was part of the same lineage that’s directly ancestral to modern East Asian genomes.)
The Denisovans who contributed to the modern Papuan genome, however, were from a completely different population. The finding hints that Denisovans were both spread over a larger geographic area than currently known and ran into H. sapiens more often than once thought.
In other words, to continue a theme that has popped up a number of times in the past few years, the story of human evolution is looking increasingly more complex. Our family tree is not the neat and stately oak-like diagram that still graces many textbooks, with H. sapiens at the apex, of course, but rather a bush of tangled branches.
Getting Species-Specific
Right about now, I know you may be thinking “wait a minute, if H. sapiens and Denisovans and Neanderthals are all separate species, how could they interbreed? Doesn’t that contradict the very definition of a species?” Well, yes. And no.
Consider, for example, that coyotes and wolves can interbreed despite being recognized as separate species. Other examples abound in the wild, including some species of bear.
But more to the point when discussing members of the genus Homo, speciation doesn’t happen overnight: It takes thousands, even hundreds of thousands of years.
According to the best information researchers have right now, based on the fairly new field of ancient DNA analysis, two populations of an as-yet-uncertain member of Homo (because some will swear it was H. erectus, others not, let’s call them Homo X) became isolated from each other somewhere between 550,000-750,000 years ago. One of those populations of Homo X was on its way into evolving into H. sapiens. The other split again, probably 380,000-480,000 or so years ago, into what would become Neanderthals and Denisovans.
Again, these splits didn’t happen in a generation or two. These were gradual processes, with the end game being separate and distinct species that should not have been able to interbreed.
Before speciation was completed, however, the different populations met up again, got busy, and successfully procreated, shuffling DNA and tangling up the branches of the family tree.
In fact, the key point from today’s research, says lead author Sharon Browning, via email from the University of Washington, is “the fact that these multiple interbreeding events occurred strongly suggests that Neanderthals and Denisovans were not so very different from us.”
Today’s research appears in Cell.