In Search of a Universal Flu Vaccine
Posted on Categories Discover Magazine
No one wants to catch the flu, and the best line of defense is the seasonal influenza vaccine. But producing an effective annual flu shot relies on accurately predicting which flu strains are most likely to infect the population in any given season. It requires the coordination of multiple health centers around the globe as the virus travels from region to region. Once epidemiologists settle on target flu strains, vaccine production shifts into high gear; it takes approximately six months to generate the more than 150 million injectable doses necessary for the American population.
Incorrect or incomplete epidemiological forecasting can have major consequences. In 2009, while manufacturers, including MedImmune and Sanofi Pasteur, were preparing vaccines against the anticipated strains, an additional influenza strain, H1N1, emerged. The prepared vaccine didn’t protect against this unanticipated strain, causing worldwide panic and over 18,000 confirmed deaths – likely only a fraction of the true number, estimated to exceed 150,000. Better late than never, a vaccine was eventually produced against H1N1, requiring a second flu shot that year.
Given that influenza has caused the majority of pandemics over the past 100 years – including the 1918 flu that resulted in as many as 50 million deaths – we’re left with the question: Can scientists produce a “universal” vaccine capable of protecting against diverse strains of influenza, one that doesn’t require annual predictions by epidemiologists and a yearly shot for you?
Vaccines Prime the Immune System to Fight
By the 18th century, and arguably much earlier in history, it was commonly known that a survivor of smallpox would not come down with it again upon subsequent exposure. Somehow, infection conferred immunity against the disease. And people recognized that milkmaids who came into contact with cowpox-ridden cattle would similarly be protected from smallpox.
In the late 1700s, farmer Benjamin Jesty inoculated his family with cowpox, effectively immunizing them against smallpox, despite future exposure. Physician Edward Jenner then catapulted humanity into a new age of immunology when he lent scientific credence to the procedure.
So if one inoculation of cowpox or one exposure to (and survival of) smallpox confers a decade’s-worth or even lifelong immunity, why are individuals encouraged to receive the flu vaccine every year?
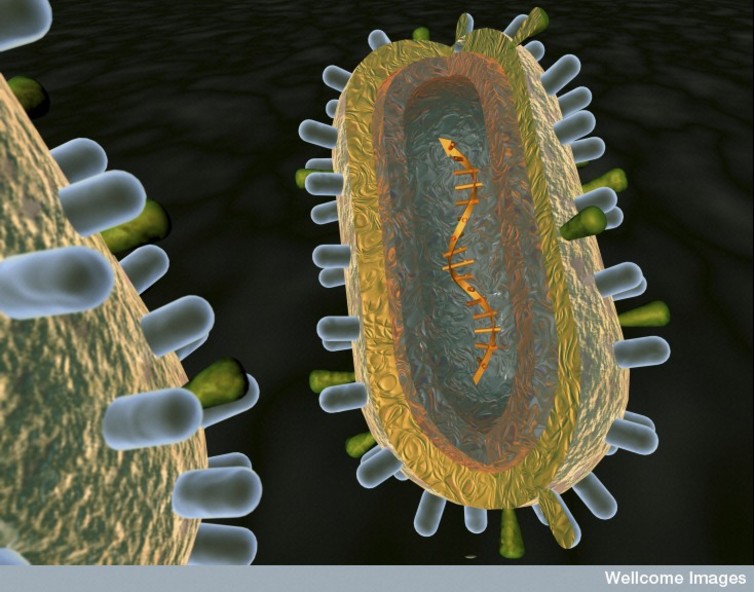
An artist’s rendition of the anatomy of a virus. (Credit: Anna Tanczos/Wellcome Images, CC BY-NC-ND)
The answer lies in how quickly the influenza virus’ anatomy changes. Each virus consists of a roughly spherical membrane encapsulating constantly mutating genetic material. This membrane is peppered with two types of “spikes”: hemagglutinin, or HA, and neuraminidase, or NA, each made up of a stem and a head. HA and NA help the virus with infection by binding to host cells, and mediate the entry of the virus into the cell and eventually its exit.
Vaccines typically elicit antibodies that target these two molecules. Once injected, an individual’s immune system gets to work. Specialized cells collect the vaccine molecules as invaders; other cells then generate antibodies that will recognize the foreign molecules. The next time those same invaders show up – whether in the form of the same vaccine or the virus strains it mimicked – the body’s immune cells recognize them and fight them off, preventing infection.
For vaccine developers, one frustrating characteristic about influenza’s mutating genome is how rapidly HA and NA change. These constant alterations are what send them back to the drawing board for novel vaccines every flu season.
Different Methods to Design a Vaccine
The smallpox vaccine was the first to use the “empirical paradigm” of vaccinology – the same strategy we largely use today. It relies on a trial-and-error approach to mimic the immunity induced by natural infection.
In other words, vaccine developers believe the body will mount an antibody response to something in the inoculation. But they don’t focus on which specific patch of the virus is causing an immune response. It doesn’t really matter if it’s a reaction to a small patch of HA that many strains share, for instance. When using an entire virus as starting material, it’s possible to get many different antibodies recognizing many different parts of the virus used in the vaccine.
The seasonal flu shot generally fits into this empirical approach. Each year, epidemiologists forecast which flu strains are most likely to infect populations, typically settling on three or four. Researchers then attenuate or inactivate these strains so they can act as the mimics in that year’s influenza vaccine without giving recipients the full-blown flu. The hope is that an individual’s immune system will respond to the vaccine by creating antibodies that target these strains; then when he or she comes into contact with the flu, the antibodies will be waiting to neutralize those strains.
But there’s a different way to design a vaccine. It’s called rational design and represents a potentially game-changing paradigm shift in vaccinology.
The goal is to design some molecule – or “immunogen” – that can cause effective antibodies to be produced without requiring exposure to the virus. Relative to current vaccines, the engineered immunogen may even allow for more specific responses, meaning the immune response targets particular parts of the virus, and greater breadth, meaning it could target multiple strains or even related viruses.
This strategy works to target specific epitopes, or patches of the virus. Since antibodies work by recognizing structures, the designers want to emphasize to the immune system the structural properties of the immunogens they’ve created. Then researchers can try to design candidate vaccines with those structures in hopes they’ll provoke the immune system to produce relevant antibodies. This path might let them assemble a vaccine that elicits a more effective and efficient immune response than would be possible with the traditional trial-and-error method.
Promising headway has been made in vaccine design for respiratory syncytial virus using this new rational paradigm, but efforts are still underway to use this approach for influenza.
Toward a universal flu vaccine
In recent years, researchers have isolated a number of potent, influenza-neutralizing antibodies produced in our bodies. While the antibody response to influenza is primarily directed at the head of the HA spike, several have been found that target HA’s stem. Since the stem is more constant across viral strains than the head, this could be flu’s Achilles’ heel, and antibodies that target this region may be a good template for vaccine design.
Researchers are pursuing a number of approaches that could cause the body to produce these antibodies of interest before becoming infected. One strategy, known as nanoparticle display, involves designing a molecule that incorporates part of the virus. In the lab, scientists could attach some combination of HA and NA particles to the outside of a spherical nanoparticle that itself is capable of causing an immune response. When injected as part of a vaccine, the immune system could “see” these molecules, and with luck produce antibodies against them.
One of the major questions that needs to be answered is what exactly should be displayed on the exterior of these nanoparticles. Some strategies display various versions of full HA molecules, while others just include stems. While more data on humans need to be collected to validate these approaches, the data from animal studies using stem-only immunogens are encouraging.
With current technology, there may never be a “one and done” flu shot. And epidemiological surveillance will always be necessary. However, it is not inconceivable that we can move from a once-per-year model to a once-every-10-years approach, and we may be within just a few years of being there.
![]()
This article was originally published on The Conversation. Read the original article.